Upserting is a way of inserting data where a new entity is only created if it doesn’t exist already. If the data already exists, then it is updated by adding new properties or by changing existing ones. The term upsert is a portmanteau of update and insert.
The concepts we discuss in this post are not specific to JanusGraph and can also be used at least for other graph databases that support the Apache TinkerPop framework. We will use a concrete example from our domain of analysing malware throughout this post to illustrate the problem of getting duplicate data. This of course doesn’t mean that the problem or the techniques we’ll discuss to solve it are specific to our domain. The example should only help us to better illustrate the problem.
This article is part of a series on graph databases
In this series we take a look at how you can use a graph database in the context of malware analysis:
Why are Duplicates a Problem?
When multiple threads insert data in parallel into a database like JanusGraph, then naturally the question occurs of how duplicates can be avoided in the database. Data coming from multiple sources or simply for unrelated events that can share some data can create situations where two or more threads insert the same data in parallel.
In our case, many malware samples are analysed in parallel by different systems which includes systems for dynamic and static analysis. We represent a malware sample as a tree-like subgraph with the sample as the root and the features as the leaf nodes the analysis systems extracted for that sample.
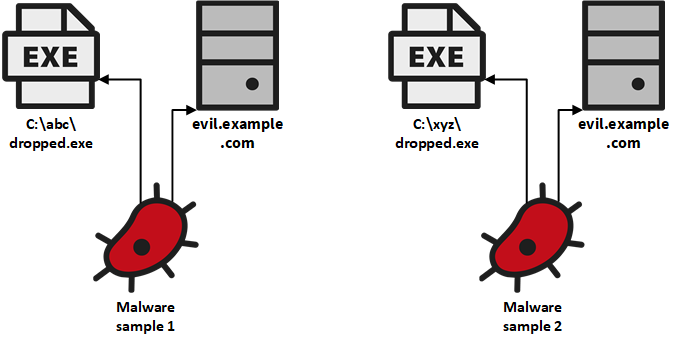
These trees are unrelated to each other before they are inserted into JanusGraph. When now two threads insert trees in parallel that share at least one feature, then we run into the problem of duplicate data in our graph. This can for example be seen for the two small trees shown in the figure above. The two malware samples contacted the same host with the domain name evil.example.com. When two threads insert these trees in parallel, then we can easily end up with two different vertices that represent the domain name. So, both samples would only have a connection to one of these domain names. This is very unfortunate for us as we couldn’t find any connection between these two samples although they clearly share a common feature, namely the domain name they contacted. That is why it is so important for us to avoid duplicates.
Using Built-In Locks
So, we have identified the problem that duplicate data prevents us from finding connections in our graph. The first solution that comes to mind is to check whether the graph database itself already offers a solution to lock data that we do not want to have duplicates of. JanusGraph can store data in different storage backends that the user can choose like Cassandra, HBase, or ScyllaDB. It uses the locking mechanism of the backend to enable uniqueness constraints which solves the problem of having duplicates in general.
However, relying on the built-in locking mechanism has the disadvantage that JanusGraph does not know our own uniqueness constraints. While it is fairly simple to define for example that only one vertex with the label Host should exist for each property value of the Name property to avoid duplicate hostnames in the graph, it becomes more complex when our services also produce duplicate edges. This can happen quite easily in our domain when for example a malware sample is analysed again, e.g., with a different configuration or in a different environment. So, we do not want to end up with multiple edges between the malware sample and the feature that we saw in both analyses runs.
Apart from not supporting more complex uniqueness constraints like these, the JanusGraph documentation also contains a warning for eventual consistent storage backends like Cassandra and in general that it can lead to deadlocks.
For those reasons, we decided not to use the built-in locking and instead enforce our uniqueness constraints outside of JanusGraph.
The Naive Approach: Get-or-Create in the Application
A very simple approach to support uniqueness constraints outside of the database is to implement them in the application that inserts the data. The most straightforward solution is to first check whether the element already exists in the graph and only insert it otherwise:
if (g.V().Has("Host", "Name", "evil.example.com").Count().Next() == 0)
{
g.AddV("Host").Property("Name", "evil.example.com").Next();
}(The code listings in this post are written in C# with Gremlin.Net but the code is basically the same in other languages for which a Gremlin language variant exists, like Java, Groovy, Python, or JavaScript.)
This simple approach already avoids most occurrences of duplicates, but it comes with an obvious risk of running into race conditions when multiple threads execute this code at the same time. When both threads execute the existence check before the first successfully added the vertex, then both threads will assume that the vertex does not exist already and therefore add it to the graph. This means that we end up with duplicates again for data that is modified at roughly the same time.
Get-or-Create Traversal
Upserting data is a use-case that is so common that a pattern emerged in the TinkerPop community to implement them in a single graph traversal. This pattern basically includes the if-check with the conditional vertex addition step in a graph traversal that does both:
g.V().Has("Host", "Name", "evil.example.com").Fold().
Coalesce<Vertex>(
Unfold<Vertex>(),
AddV("Host").Property("Name", "evil.example.com")).
Next();This traversal returns the vertex, either as an existing one retrieved with the has step or a newly added one, which allows adding more properties or edges. Explaining the traversal itself in greater detail is beyond the scope of this blog post but you can read a nice explanation in this StackOverflow answer or in the freely available book Practical Gremlin (which is a very good resource to learn Gremlin in general).
By using this traversal, we avoid the network round-trips between the existence check and the conditional vertex creation we had before in our application. However, the traversal will usually not be executed as an atomic operation which means that still some time passes between the existence check and the vertex addition. So, the traversal does not completely resolve the race conditions. It just reduces the time window a lot due to the omitted additional network round-trip between our application and the JanusGraph Server.
This small time window – which should be usually in the range of a few milliseconds, depending on the concrete setup – may be small enough for many scenarios. Unfortunately for us, we still encountered a noticeable number of duplicates, especially for vertices that occur frequently in our analysis systems. For example, the domain name google.com led to duplicate vertices as it is frequently used by malware to perform internet connectivity checks. That is why we added a distributed locking mechanism in addition to this upsert traversal.
Add Distributed Locking
When just a single process is responsible for upserting data into the database, then a simple lock around the upsert traversal in the application might be a good solution. But what’s the point of using a distributed database when we then restrict ourselves to insert data into it from only a single process (which of course also means from a single application)?
That is why we decided to use a distributed locking algorithm. This distributed locking enables us to ensure that only one process can upsert a particular vertex or edge at the same time. We do this by locking individual vertices or edges by the properties that make them unique for us. The locking can of course significantly slow down upserts, especially when some vertices are very frequent and when waiting for their lock blocks the upsert of a whole subgraph (e.g., of a malware sample with its observed features like in our use case). This performance impact is small enough in our case to not be an issue.
There exist various algorithms that implement distributed locking. We use the Redlock algorithm which is based on the Redis in-memory database because we already had some experiences with Redis and because libraries for various languages exist that implement this algorithm. It should however be noted that the Redlock algorithm can only guarantee that a lock is held by just one process at a time under certain assumptions. Clock drifts or delayed network problems can for example violate those assumptions. You can read more about the assumptions and in which cases they can be violated in this analysis of the Redlock algorithm.
JanusGraph Specific Settings
There are a few settings in JanusGraph that should be considered in order to avoid duplicates.
First of all, database level caching should be disabled as it easily leads to duplicate edges when more than one JanusGraph instance performs the upserts. The reason for this is that this cache includes vertices with their adjacency lists. When an edge should be upserted into the graph, then the get-or-create traversal would not find an edge that was already added to the vertex by another JanusGraph instance after the vertex was added to the cache. Note that the database level cache is already disabled by default in JanusGraph.
For storage backends that allow configuration of the consistency level, strong consistency should be chosen. In the case of Cassandra or ScyllaDB, QUORUM can be used to ensure strong consistency which is also already the default setting of the JanusGraph CQL backend.
Apart from these settings, we want to avoid the built-in locks of JanusGraph and therefore all schema settings should be avoided that lead to locks. This includes not only property uniqueness but also all other edge label multiplicities than MULTI.
Outlook: Native Upsert Functionality in JanusGraph?
The distributed locking mechanism discussed earlier is of course only a workaround for a missing functionality in JanusGraph. If JanusGraph supported upserts, then we wouldn’t have to implement locking outside of it. To support upserts directly, JanusGraph needs a backend that provides serializable isolation and atomic operations across different rows. These are two of the four ACID properties. Serializable isolation means that transactions have the same effect as if they were executed serially one after another which avoids the race condition between different get-or-create traversals that leads to duplicates. Atomic operations could on top of that ensure that either all changes from a traversal are written to the database or no changes in case of an error. This needs to be supported across rows for edges because JanusGraph stores edges as an adjacency list in the rows of both vertices that are connected by the edge.
Currently, the Berkeley DB storage backend is the only official JanusGraph backend that supports these properties, but this backend is mainly used for testing purposes or for relatively small graphs. Distributed databases like Cassandra or HBase traditionally don’t support these transaction properties. FoundationDB however is a distributed database that supports ACID transactions and Ted Wilmes, a founding member of JanusGraph, developed a first version of a storage adapter for FoundationDB. This storage adapter can provide a way for JanusGraph to support upserts without losing its advantage of being a horizontally scalable graph database.
Conclusion
Avoiding duplicates in a graph database is a frequent requirement. The get-or-create Gremlin traversal is a good way to ensure that new entities are only created if they do not already exist in the graph. However, it is not an atomic operation and therefore comes with a race condition where duplicates still occur due to multiple upserts of the same entity at roughly the same time. Adding a dedicated distributed locking mechanism can solve this race condition and thereby enable upserts in a reliable way with a graph database like JanusGraph that currently does not support them directly. The new FoundationDB storage adapter can provide a way forward for JanusGraph to add this support which would make the locking unnecessary.



